Example Using GUSB at UCSC
Searching for similar sequences of GUSB exons, in human genome,
using BLAT on the UCSC server returns this result (extract):
Lets load this file in
NumericalFISH (using all default values):
We can easily see that the result of the BLAT query are much easier
to interpret:
- The 12 exons of the gene are retrieved on strand + of
chromosome 7
- There is a dupplication of this gene on the minus strand just
in front, but some exons seem to be missing
- There is another partial dupplication of some exons on the
strand + of the same crhomosome
- There are a large number of dupplications (with losses of
exons) on chromosome 5
- There are also dupplication on chromosome 6
- Some exons seem to be retrieved on a number of chromosomes
Lets double click on chromosome 7
We now have a very clear view of
the duplication event on this chromosome.
Lets mouse over the first exon of
the gene (1)
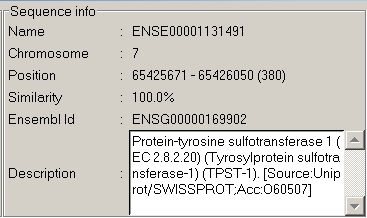
Then we mouse over the copy of exon
12 located near the top of the chromosome (2):
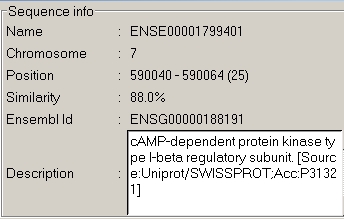
In both cases, information is
available from Ensembl, and these two domains are recognized as a
known gene.
We can now change the numbers of
chromosomes per line and the zoom to see:
And we can save the image (under jpeg here) for further work with an
image dedicated software (the Gimp ?):
Back to home






