Use for example this fasta
file (several exons from GUSB)
. BLAT
it on the human genome (since these are exons from a human gene).
Be carefull ! Exons downloaded from ENSEMBL have gene ID (here
ENST00000304895) as first word and this while return an error
(duplicated ids).

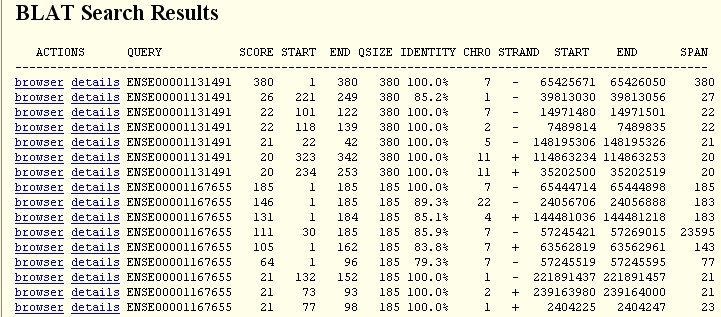
As shown above, the raw result is not very easy to analyze
quickly.
Simply save the resulting html file as for example SCL_exons_hgBlat.htm.
Set the SearchTool to BLAT.
Use for example this fasta
file (several exons from GUSB)
. BLAT
it on the human genome (since these are exons from a human gene).
Now on this server this can be a bit complicated, because each
exon is treated separately, as shown below:
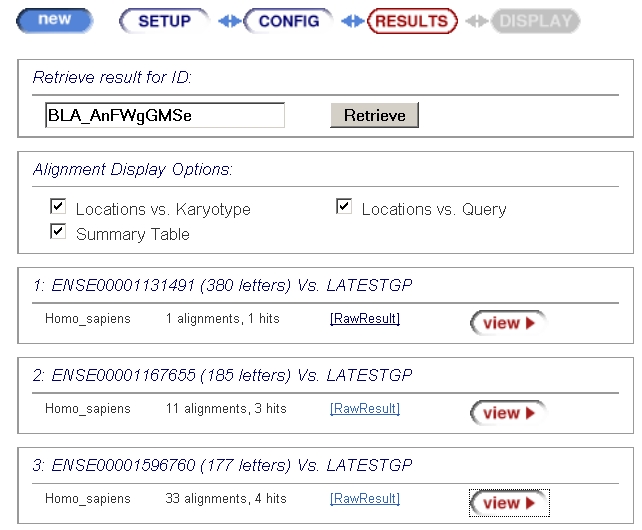
Now you need to successively hit each "RawResult", and save each
page under a different name.
Download the program from UCSC, install your database on a local port. Run it as detailled. Use the output file as you would from UCSC web server, except this will be a text file and not an html file.
The procedure is exactly identical (but the response is much longer to obtain !).
Download the program from the NCBI ftp
site. Format your database and run your blastn. Then the procedure
is identical to that used for EBI, except that all results
will be in a single file.
Make sure you use the "-m 7" option, that create a XML
ouput file. Save it with an extension like ".txt", ".out" or
".BLAST".